Simulate some fake data
dat <- gamSim(4, n = 400, dist = "normal", scale = 1)## Factor `by' variable exampleThe simulation creates a dataframe with a single response and 3 numeric covariates. The effect of
each covariate depends on the value of a factor, fac.
We can take a look at the data by plotting the response against each of the covariates:
ggplot(dat, aes(x = f1, y = y)) +
geom_point(size = 1.5) +
ggtitle("y versus f1:") +
facet_wrap( ~ fac)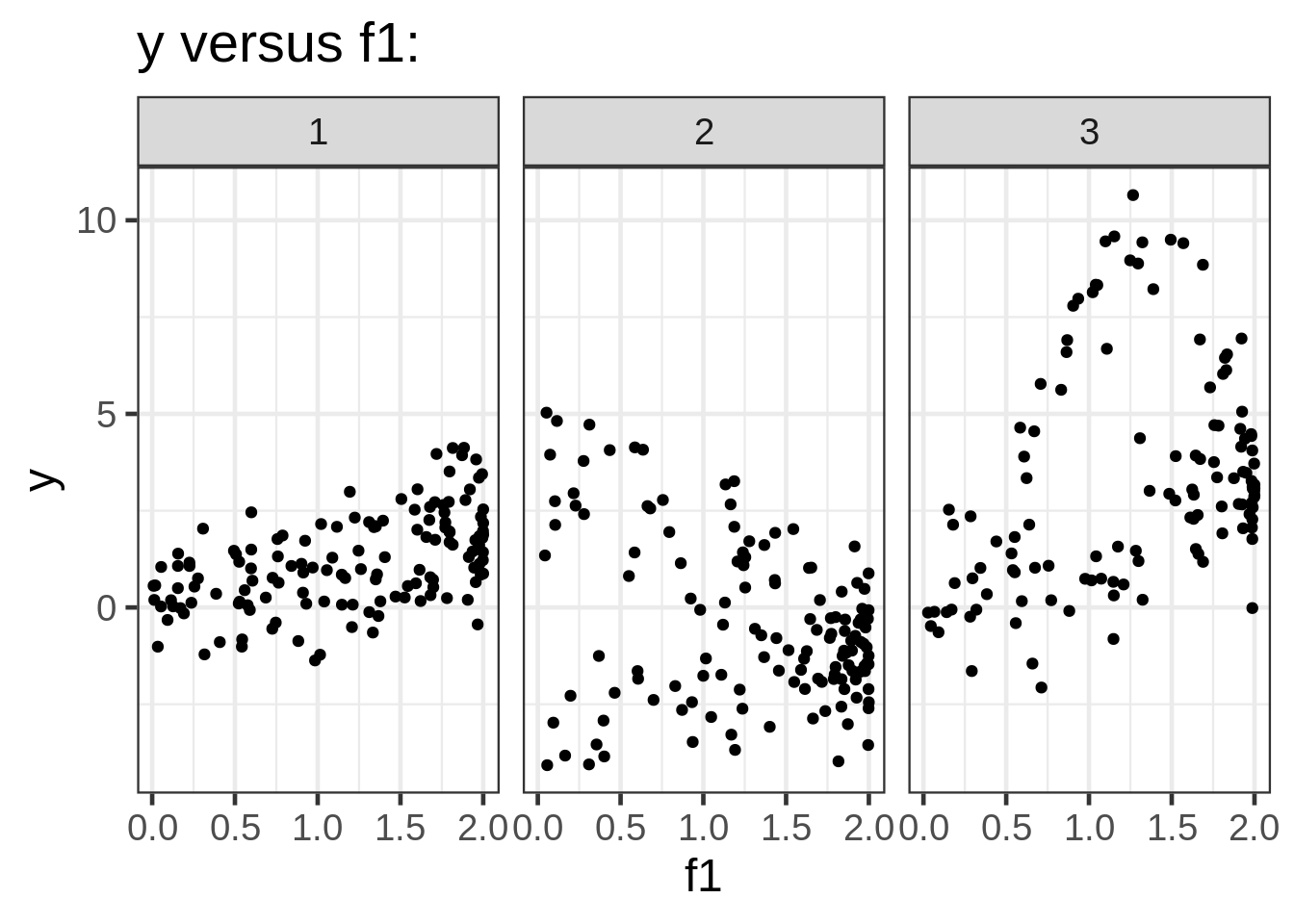
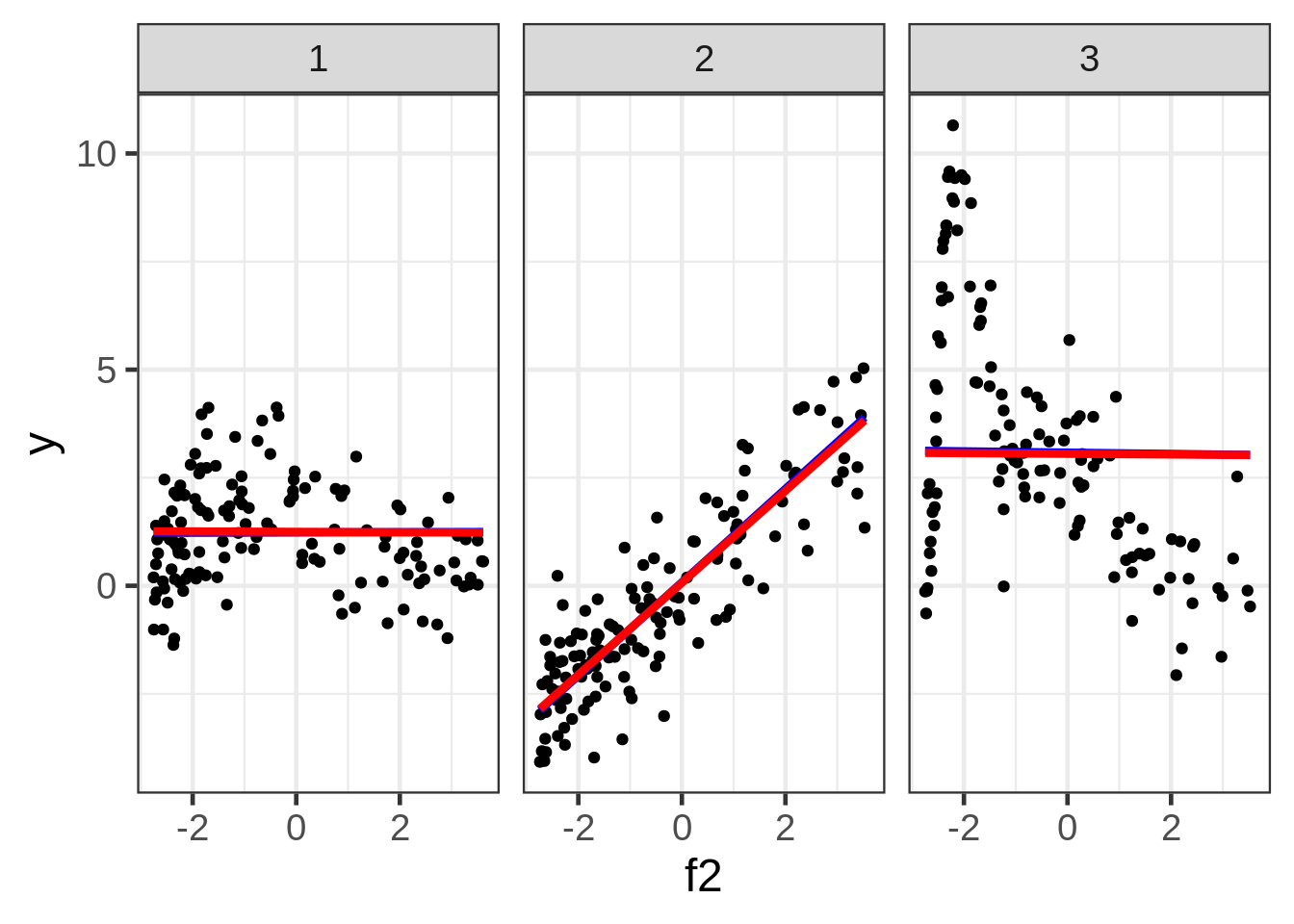
ggplot(dat, aes(x = f2, y = y)) +
geom_point(size = 1.5) +
ggtitle("y versus f2:") +
facet_wrap( ~ fac)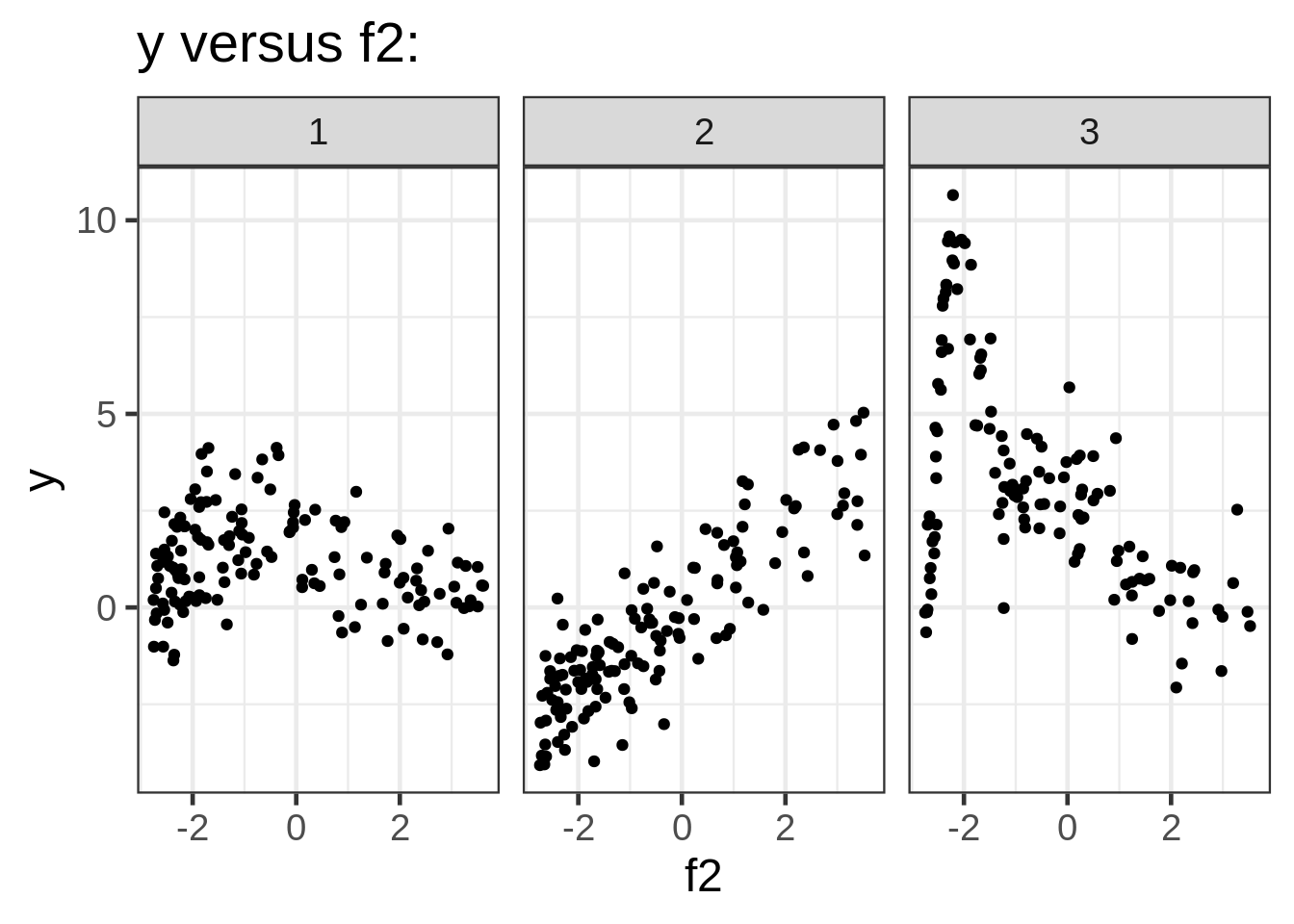
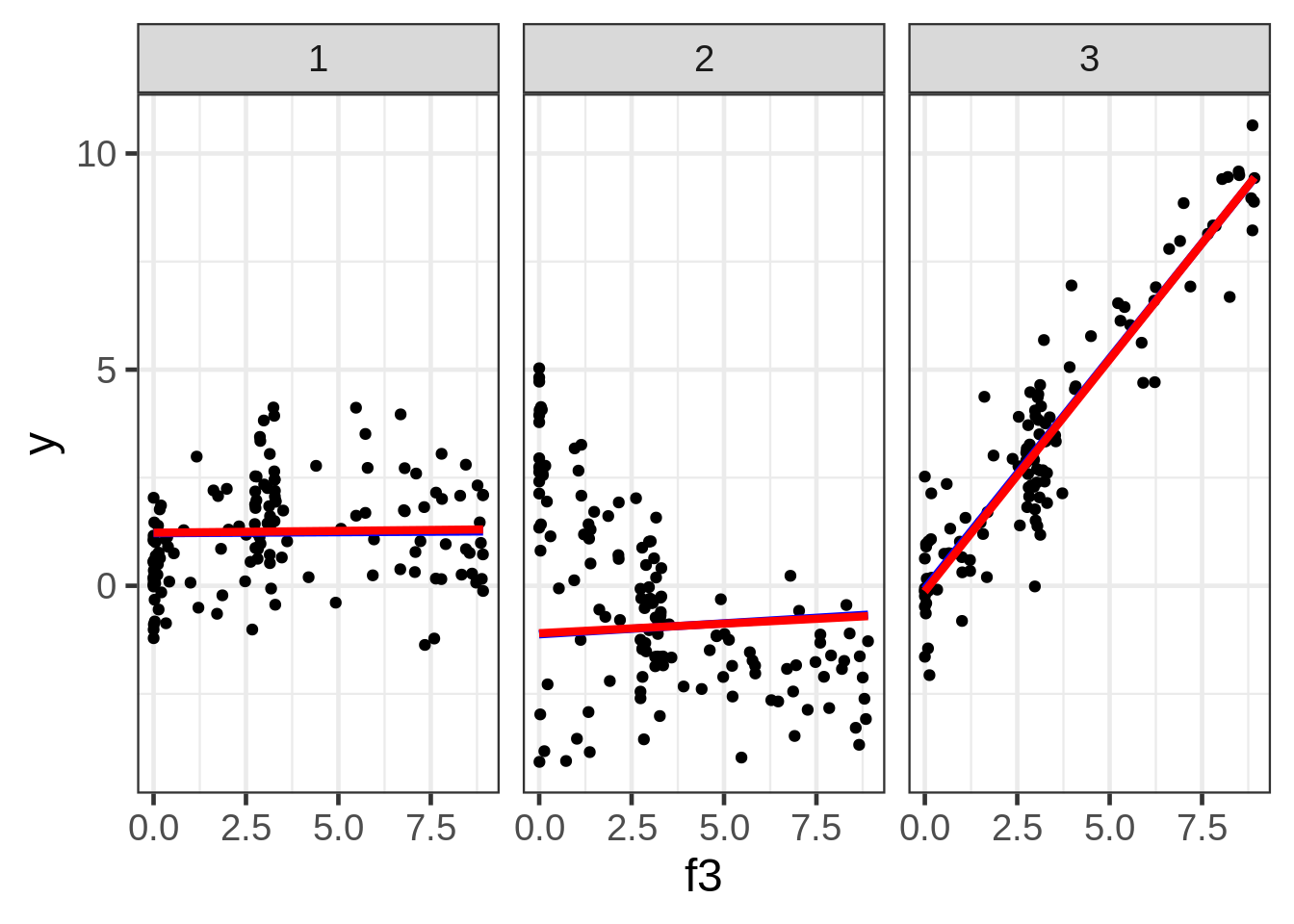
ggplot(dat, aes(x = f3, y = y)) +
geom_point(size = 1.5) +
ggtitle("y versus f3:") +
facet_wrap( ~ fac)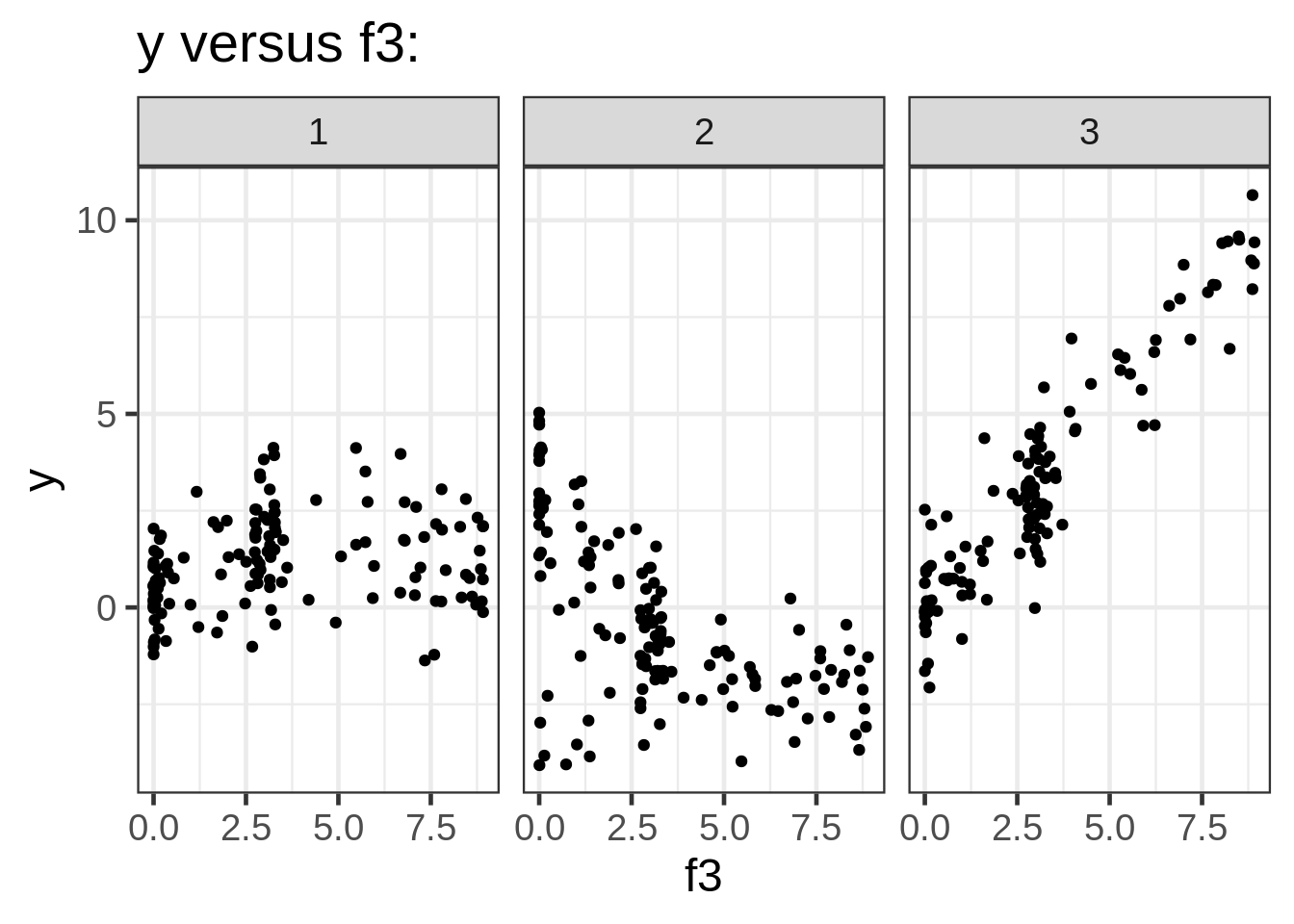
Separate Smoothers
The usual way to fit the data in mgcv is to use the by argument. This results in a different
smooth for each level of the factor. Ie, the smoothing parameters for each of the continuous factors
is allowed to vary with each level of fac. Now, we can get really fancy here by changing the
knots, smoothers, adding interactions, model checking, yada yada, but I just want a model fit to
compare apples-to-apples with the other method.
fit1 <- gam(y ~ fac + s(f1, by = fac) + s(f2, by = fac) + s(f3, by = fac), data = dat)
dat$preds1 <- predict(fit1)A summary of the model fit shows that separate smooths are created for each numeric covariate and
each level of fac:
summary(fit1)##
## Family: gaussian
## Link function: identity
##
## Formula:
## y ~ fac + s(f1, by = fac) + s(f2, by = fac) + s(f3, by = fac)
##
## Parametric coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.23062 0.08879 13.86 <2e-16 ***
## fac2 -1.69374 0.12581 -13.46 <2e-16 ***
## fac3 2.21761 0.12962 17.11 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Approximate significance of smooth terms:
## edf Ref.df F p-value
## s(f1):fac1 1.703 2.129 19.367 4.83e-09 ***
## s(f1):fac2 1.629 2.019 0.767 0.490
## s(f1):fac3 1.000 1.000 0.274 0.601
## s(f2):fac1 1.000 1.000 0.003 0.957
## s(f2):fac2 1.000 1.000 233.375 < 2e-16 ***
## s(f2):fac3 1.000 1.000 0.048 0.827
## s(f3):fac1 1.000 1.000 0.009 0.925
## s(f3):fac2 1.000 1.000 0.939 0.333
## s(f3):fac3 1.000 1.000 435.018 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## R-sq.(adj) = 0.843 Deviance explained = 84.8%
## GCV = 1.1138 Scale est. = 1.0767 n = 400ggplot(dat, aes(x = f1, y = y)) +
geom_point(size = 1.5) +
geom_line(aes(y = preds1), size=1.5, color = "blue") +
facet_wrap( ~ fac)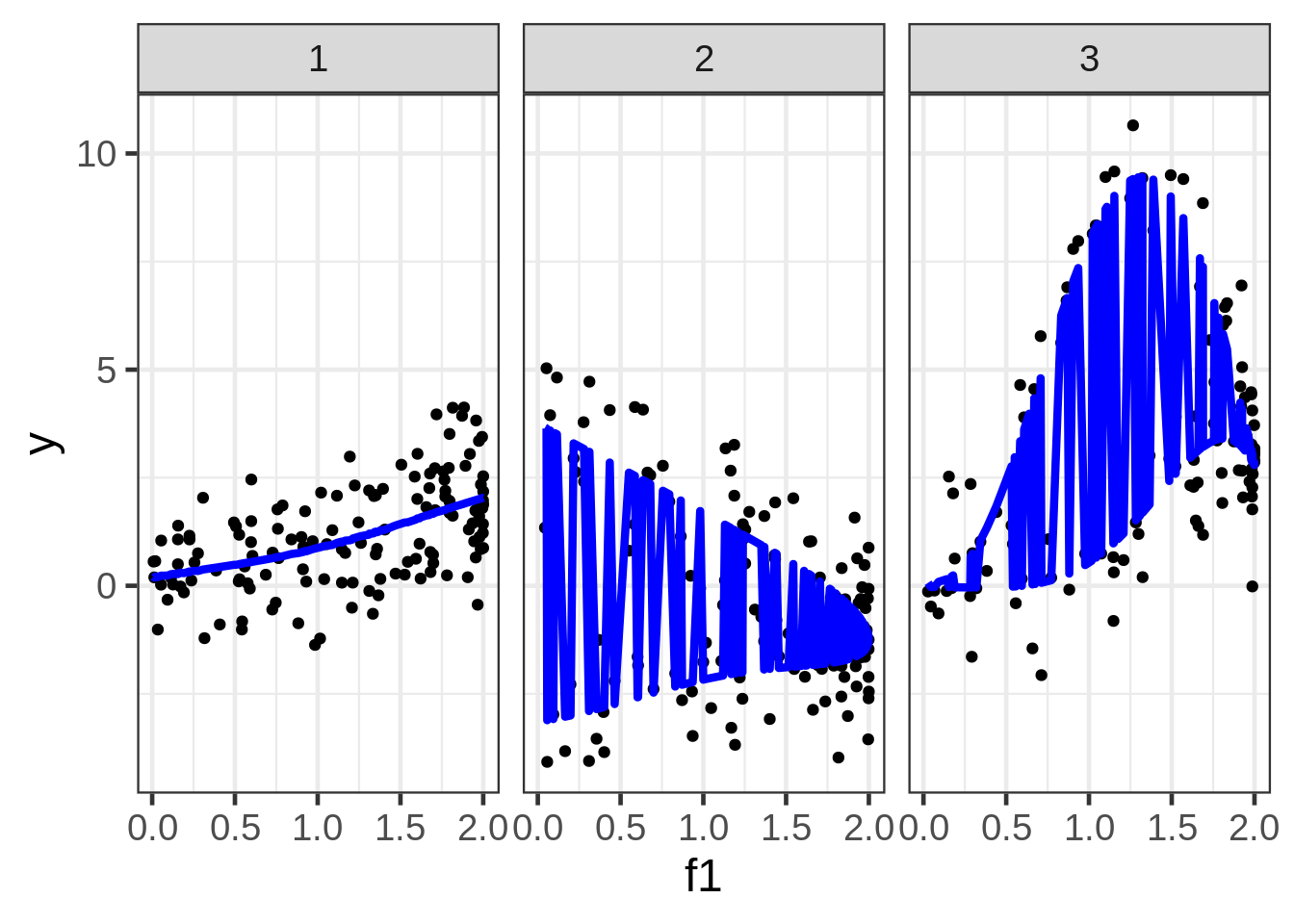
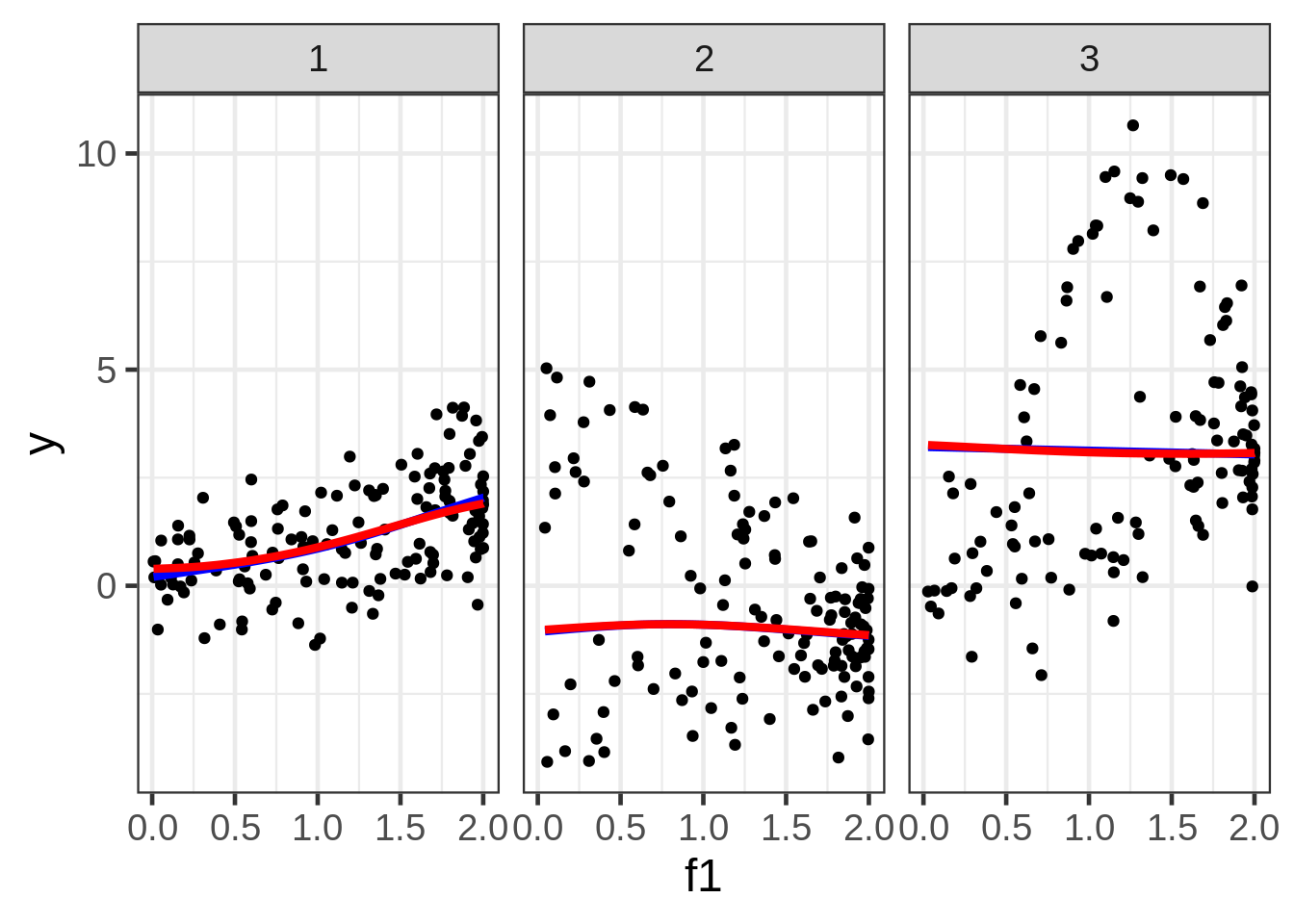
We can plot the predictions, but the picture is very poor because the preds also depend on f1 and f2, which are not shown, and are varying behind the scenes. Although this gives us some nice information about the extent that \(E(y)\) could vary as a function of all three covariates, it’s almost certainly not the plot we want to draw. A solution is to fix f2 and f3 at some representative values.
new <- data.frame(
f1 = dat$f1,
fac = dat$fac,
f2 = median(dat$f2),
f3 = median(dat$f3))
dat$preds1_f1 <- predict(fit1, newdata = new)ggplot(dat, aes(x = f1, y = y)) +
geom_point(size = 1.5) +
geom_line(aes(y = preds1_f1), size=1.5, color = "blue") +
facet_wrap( ~ fac)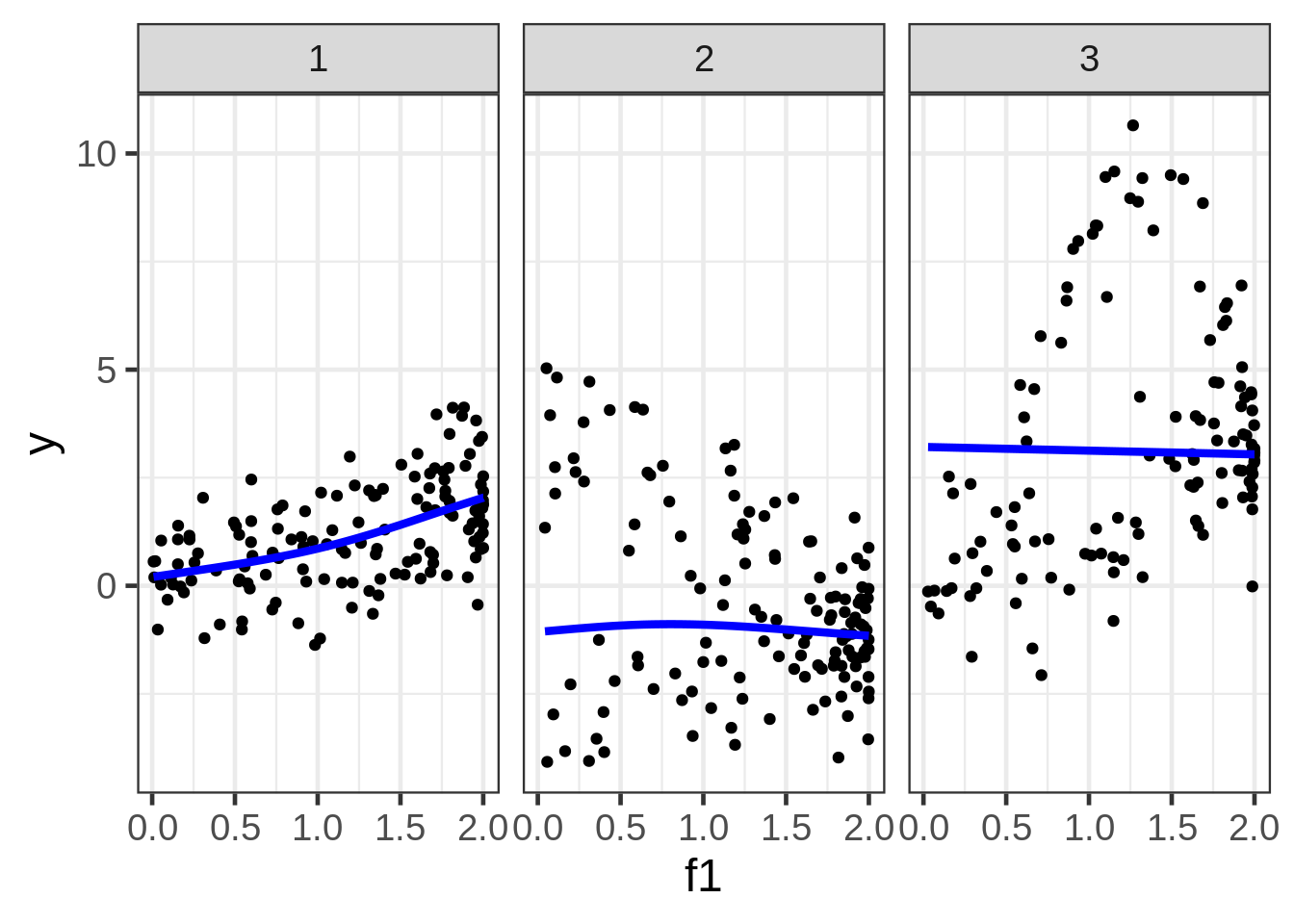
Well, the resulting marginal1 smooths are not very exciting. The smooth for second facet looks particularly misleading, but I don’t see a way around that when we are in the business of making a 2D plot of what is a 5D relationship.
I use the word ‘marginal’ here in the sense of an estimated marginal mean. Really, ‘conditional smooth’ is the more proper terminology.↩